About Me
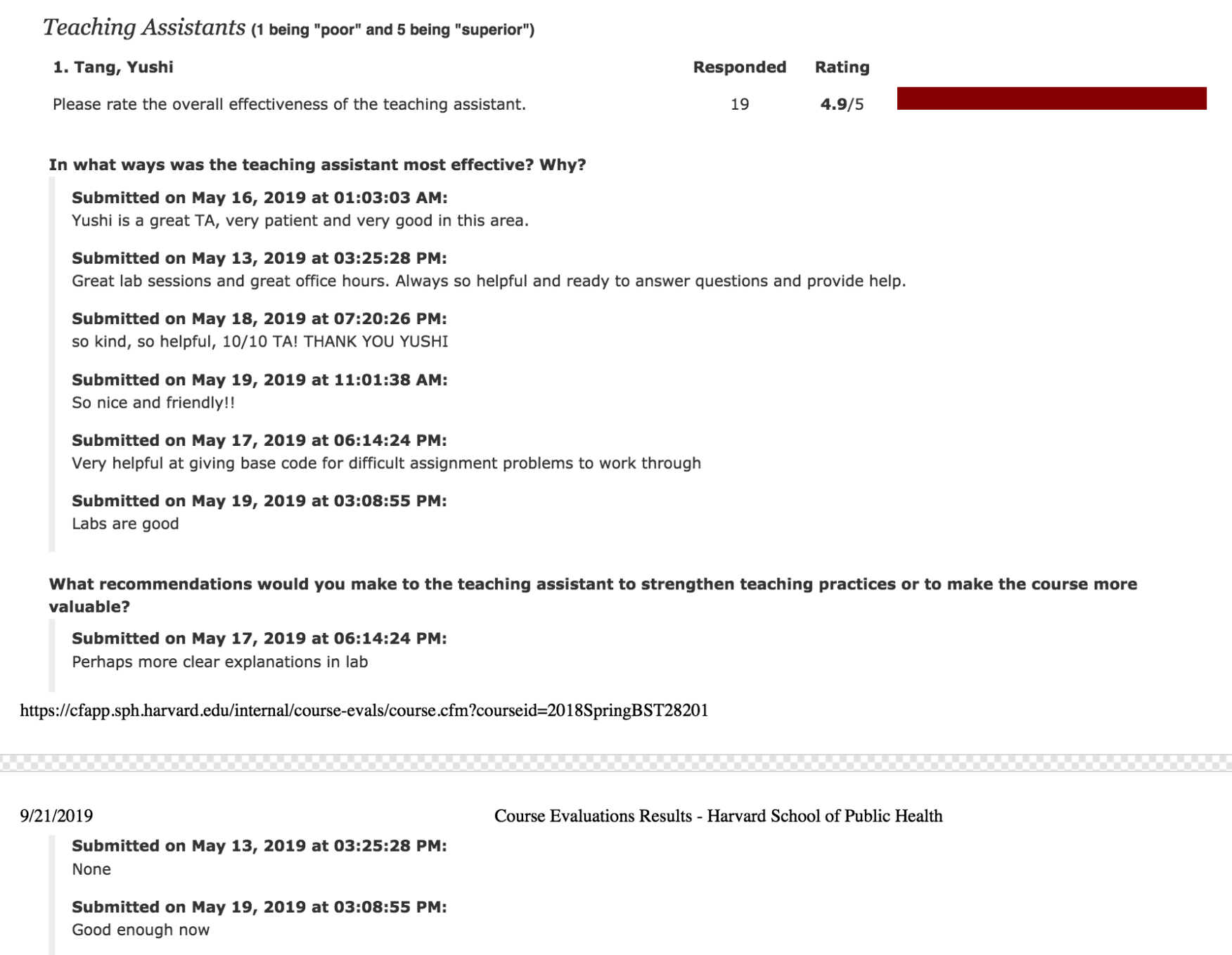
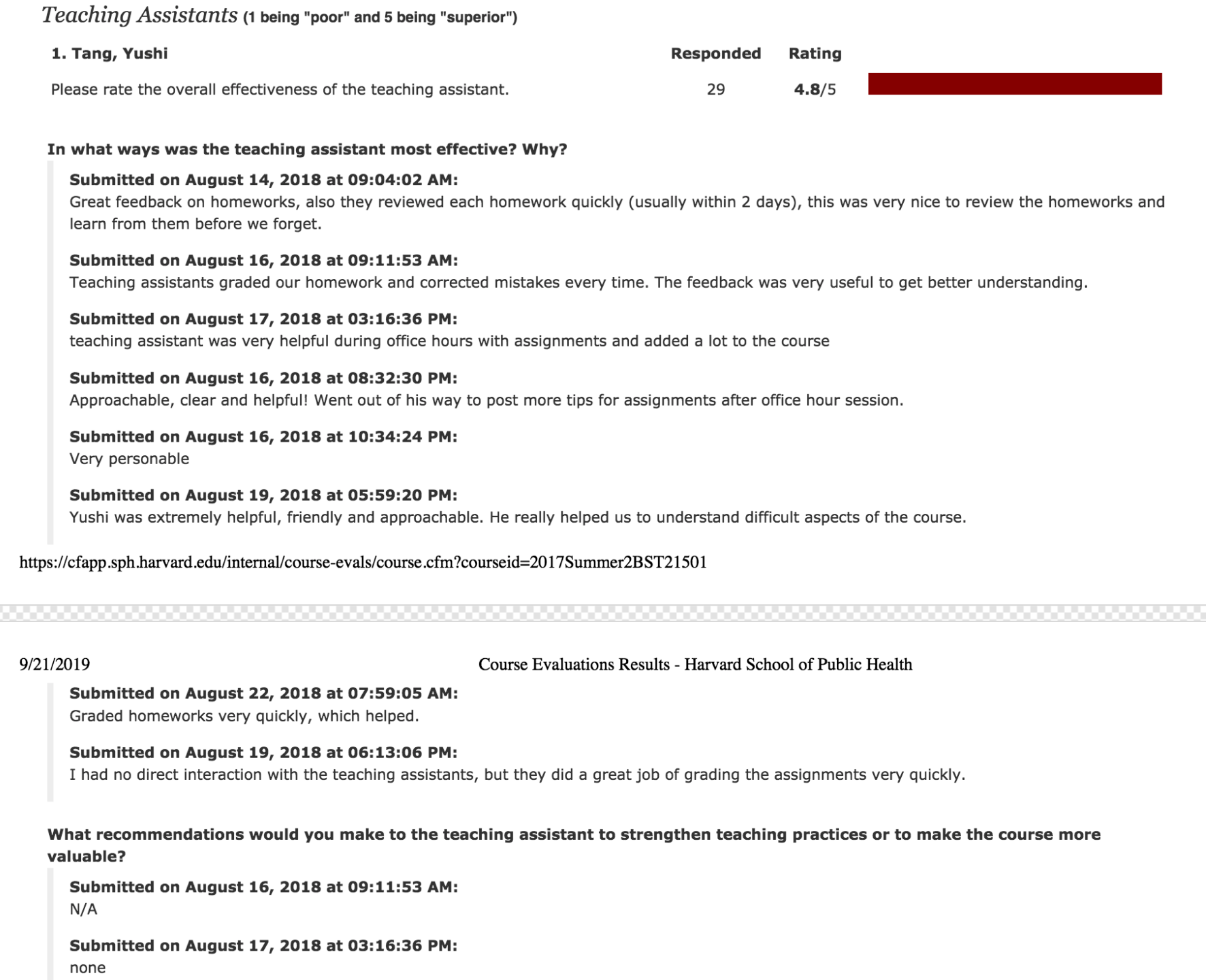
I am a biostatistics researcher at the Storey Lab for Statistical Genomics in the Lewis-Sigler Institute for Integrative Genomics, Princeton University.

I hold a Ph.D. degree in Quantitative and Computational Biology at Princeton University and a Graduate Certificate in Statistics and Machine Learning at Princeton CSML.
Prior to joining Princeton, I received my Bachelor of Science and Bachelor of Economics from Peking University in July 2017, received my Master of Science from Department of Biostatistics at Harvard University in May 2019.
Research Interest
I develop causal inference and statistical methods with applications in genetics and genomics. My recent work focuses on the following areas.
Methodology
Applications
Software
Awards and Honors
- Reviewers’ Choice Abstract Award (Top 10%), awarded by the American Society of Human Genetics, 2022
- The King Peh Kwoh Fellowship awarded by Princeton University, 2020
- Lewis-Sigler Institute Scholars Award in Quantitative and Computational Biology, awarded by Lewis-Sigler Institute, Princeton University, 2019
- Outstanding Graduate of Beijing as a Bachelor, awarded by the Beijing Municipal Commission of Education, 2017
- Excellent Graduate who has demonstrated outstanding performance, awarded by Peking University, 2017
- UCLA Cross-disciplinary Scholars in Science and Technology Award in recognition of outstanding research and presentation skills, 2016
- Tang Xiaoyan Scholarship for innovation in environmental science, awarded by College of Environmental Sciences and Engineering, Peking University, 2016
- National Scholarship (Top 0.2%), awarded twice by the Ministry of Education of China, 2016 & 2015
- Pacemaker to Merit Student (Top 1%) in recognition of outstanding academic performance, awarded twice by Peking University, 2016 & 2015
- May Fourth Scholarship (Top 10%) in recognition of excellent academic performance, awarded by Peking University, 2014
Publications
Note: [J] for journals, [P] for preprints, [C] for conference proceedings.
[1] Tang Y, Cabreros I, Storey JD. (2026) Identifying causal genotype-phenotype relationships for population-sampled parent-child trios [J]. Genetic Epidemiology 50(1): e70027. DOI:10.1002/gepi.70027
[2] Tang Y and Storey JD. (2025) A generalized test of genotype-phenotype causality in population-sampled nuclear families [P]. bioRxiv. DOI:10.64898/2025.12.29.696865
[3] Tang Y, Cabreros I, Storey JD. (2024) Identifying causal genotype-phenotype relationships for population-sampled parent-child trios [P]. bioRxiv. DOI:10.1101/2024.12.10.627752
[4] Xiong F, Su Z, Tang Y, Dai T, Wen D. (2024) Global WWTP Microbiome-based Integrative Information Platform: From experience to intelligence [J]. Environmental Science and Ecotechnology 20: 100370. DOI:10.1016/j.ese.2023.100370
[5] Li F, Bao Y, Chen L, Su Z, Tang Y, and Wen D. (2023) Screening of priority antibiotics in Chinese seawater based on the persistence, bioaccumulation, toxicity and resistance [J]. Environment International 179: 108140. DOI:10.1016/j.envint.2023.108140
[6] Su Z, Wen D, Gu AZ, Zheng Y, Tang Y, and Chen L. (2023) Industrial effluents boosted antibiotic resistome risk in coastal environments [J]. Environment International 171: 107714. DOI:10.1016/j.envint.2022.107714
[7] Tang Y and Storey JD. (2022) On the polygenic trait model in population-based human genetics studies: What is random and what is fixed [C]. The 72nd Annual Meeting of The American Society of Human Genetics. 2022 Sep. Reviewer’s Choice Abstract Award (PDF)
[8] Tang Y, Dai T, Su Z, Hasegawa K, Tian J, Chen L, and Wen D. (2020) A tripartite microbial-environment network indicates how crucial microbes influence the microbial community ecology [J]. Microbial Ecology 79: 342–356. DOI:10.1007/s00248-019-01421-8
[9] Zhao M, Tang Y, Kim H, and Hasegawa K. (2018) Machine learning with k-means dimensional reduction for predicting survival outcomes in patients with breast cancer [J]. Cancer Informatics 17: 1176935118810215. DOI:10.1177/1176935118810215
[10] Dai T, Zhang Y, Ning D, Su Z, Tang Y, Huang B, Mu Q, and Wen D. (2018) Dynamics of sediment microbial functional capacity and community interaction networks in an urbanized coastal estuary [J]. Frontiers in Microbiology 9: 2731. DOI:10.3389/fmicb.2018.02731
[11] Su Z, Dai T, Tang Y, Tao Y, Huang B, Mu Q, and Wen D. (2018) Sediment bacterial community structures and their predicted functions implied the impacts from natural processes and anthropogenic activities in coastal area [J]. Marine Pollution Bulletin 131: 481–495. DOI:10.1016/J.MARPOLBUL.2018.04.052
[12] Dai T, Zhang Y, Tang Y, Bai Y, Tao, Y, Huang B, and Wen D. (2016) Identifying the key taxonomic categories that characterize microbial community diversity using full-scale classification: a case study of microbial community in the sediments of Hangzhou Bay [J]. FEMS Microbiology Ecology 92(10): fiw150. DOI:10.1093/femsec/fiw150